
Blog
Mining wastewater data to refine COVID-19 case estimation

September 11, 2020
Why it matters
The presence of SARS-CoV-2 in the feces of COVID-19 patients and in wastewater has drawn attention to the use of wastewater surveillance as an epidemiological tool. Communities are looking at SARS-CoV-2 viral titers to understand trends over time, and further inform reopening plans for states and school systems. However, we believe that there’s much more that can be learned from wastewater data. For example, at Biobot we have been pioneering the development of models to estimate the number of COVID-19 cases from viral titers. Estimating cases from wastewater provides useful complementary data to clinical testing results, because wastewater reflects all individuals regardless of their symptoms or access to testing.
Our approach
At Biobot Analytics, we are working with hundreds of communities across the nation and we have generated the world’s largest COVID-19 wastewater dataset. Since we first successfully quantified SARS-CoV-2 in wastewater in March 2020, we have analyzed nearly 3,000 wastewater samples from over 400 locations in the US, with over 60% testing positive for the SARS-CoV-2 virus. This extensive database is the basis for the case estimation model presented here.
When we compare the total virus load in each of these wastewater samples with the number of reported COVID-19 cases in the respective communities, the amount of SARS-CoV-2 virus observed in the sewer system correlates strongly with the average number of individuals who tested positive for COVID-19 in 1–7 days following the wastewater sampling. This empirical relationship is consistent with our previous finding that wastewater is a leading indicator of COVID-19 cases, and helps us estimate the number of COVID-19 cases from wastewater. By mining this large and unbiased historical dataset, we can model the relationship between the amount of virus in sewage and the associated number of reported cases.
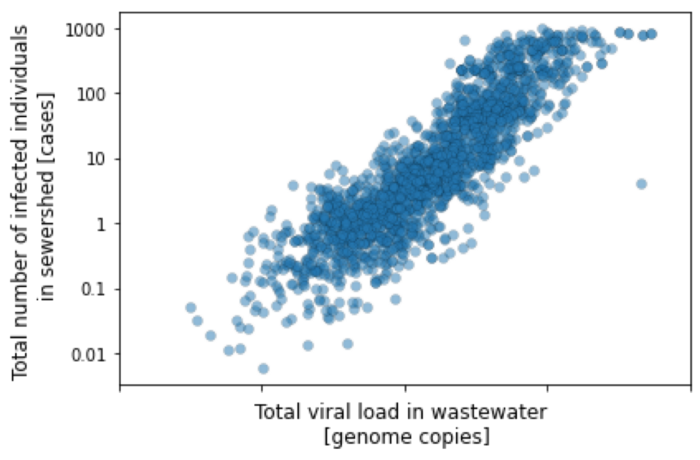
Using this empirically-derived relationship aligns with the standard approach used to estimate cases in the field of wastewater-based epidemiology. The conventional approach for estimating cases from wastewater samples is to divide the total amount of virus in wastewater by the amount of virus shed per infected person.
However, this conventional approach is dependent on a virus shedding parameter obtained from clinical studies, which is extremely difficult to reliably measure. The clinical studies used to develop this parameter are often limited to a small number of individuals, measure viral load in patients who are well into the course of their infection, and usually study more severe cases of COVID-19 that result in hospitalization. Because of this, the results of these studies vary widely, with viral loads ranging over multiple orders of magnitude. Moreover, recent studies have shown that early, pre-symptomatic infection is accompanied by a large burst of viral shedding, orders of magnitude higher than reported in clinical studies. This means that the clinical approach of obtaining the true virus shedding parameter would require testing COVID-19 patients before they develop symptoms and seek out clinics. This is not feasible.
The benefit of our modeling approach is that it’s independent of clinical studies on viral shedding and less affected by person-to-person variability of virus shedding, temporal viral shedding dynamics within any one individual, and the proportion of COVID-positive people shedding the virus in their stool. This alternative data-driven approach is only possible with a large enough dataset, and will only improve as we gather more data and our testing improves. Currently, this approach is based on reported clinical cases. In the future, we’ll want to incorporate testing rates and asymptomatic populations into our estimates of infected individuals in communities.
As we start estimating cases from wastewater more reliably, wastewater surveillance becomes a more potent public health tool, complementing costly in-person COVID testing. Moreover, because wastewater data is an indicator of near-future cases, we see this becoming an invaluable tool in guiding proactive public health responses.
References
Wu, F. et al. (2020) SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. https://msystems.asm.org/content/5/4/e00614-20
He, X et al. (2020) Temporal dynamics in viral shedding and transmissibility of COVID-19. Nature Medicine. https://www.nature.com/articles/s41591-020-0869-5
Wu, F. et al. (2020) SARS-CoV-2 titers in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. medRxiv. https://www.medrxiv.org/content/10.1101/2020.06.15.20117747v2
Written by Dr. Noriko Endo
Noriko is a Research Program Manager at Biobot and has been working at the intersection of engineering and public health for over 8 years. In her spare time, Noriko is either playing ice hockey or cheering on the Boston Red Sox.





