
Blog
Using Genomic Sequencing to Monitor COVID Variants and Subvariants in Wastewater

June 27, 2022
Throughout the COVID-19 pandemic, wastewater-based epidemiology has been a reliable independent indicator of viral spread, and has become an even more important leading signal as clinical case reporting lags. But is wastewater also useful for tracking new variant activity? The short answer is yes!
How can we monitor variants with wastewater testing?
In general, there are two ways by which we can identify SARS-CoV-2 variants in wastewater:
- Develop new qPCR assays for each variant, or
- Use viral genomic sequencing
The best method for monitoring variants in wastewater
Although running genomic sequencing takes time (getting results can take up to two weeks), viral genomic sequencing is preferable because once developed, the test doesn’t need to be changed in order to identify new variants. This is incredibly important as we’ve seen variants sweep through communities in mere weeks, often quicker than a new qPCR variant-specific assay can be developed.
Unlike qPCR, genomic sequencing does not quantify the amount of SARS-CoV-2 present in the sample, but can determine the relative percentages of each variant and even subvariants present in a given sample. This can be used to track variants over time as new ones increase from a small percentage to a larger percentage within communities.
Tracking the rise of Omicron in wastewater
During the Delta variant surge, our team built a bioinformatics pipeline to identify variants from the sequencing data, creating a versatile platform that can pivot quickly to detect new variants in wastewater.
Based on our sequencing data, we saw the Omicron variant rapidly overtake Delta in December, 2021. Initially appearing in early December, by the end of month, Omicron represented the majority of circulating SARS-CoV-2 in wastewater.
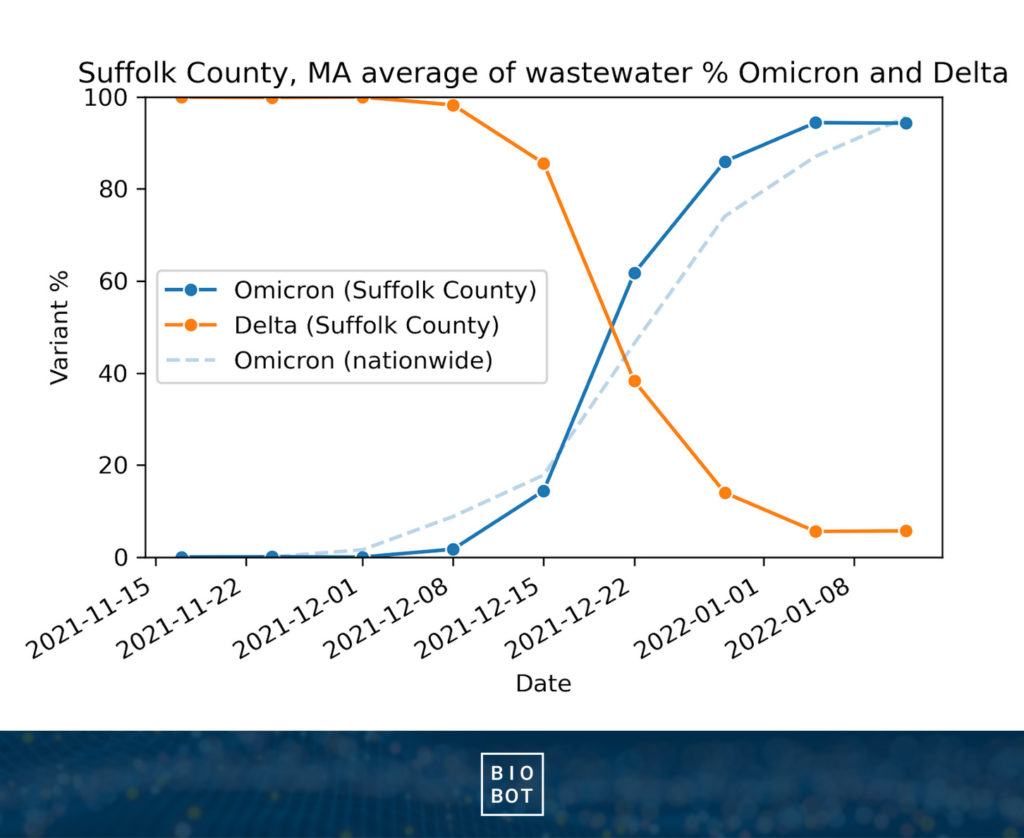
Notably, when aggregated at the national level, the transition between Delta and Omicron appears more gradual. However, when you look at specific locations (ie. Suffolk County), you can see just how quickly Omicron took over at the community level. In most locations across the US, Omicron became dominant in only two weeks, taking about a month to become the most prevalent variant across the US.
Tracking Omicron subvariants in wastewater
Over the past four months, we’ve seen Omicron subvariant BA.2/2.12 slowly climb, and eventually take over as the major subvariant nationwide.
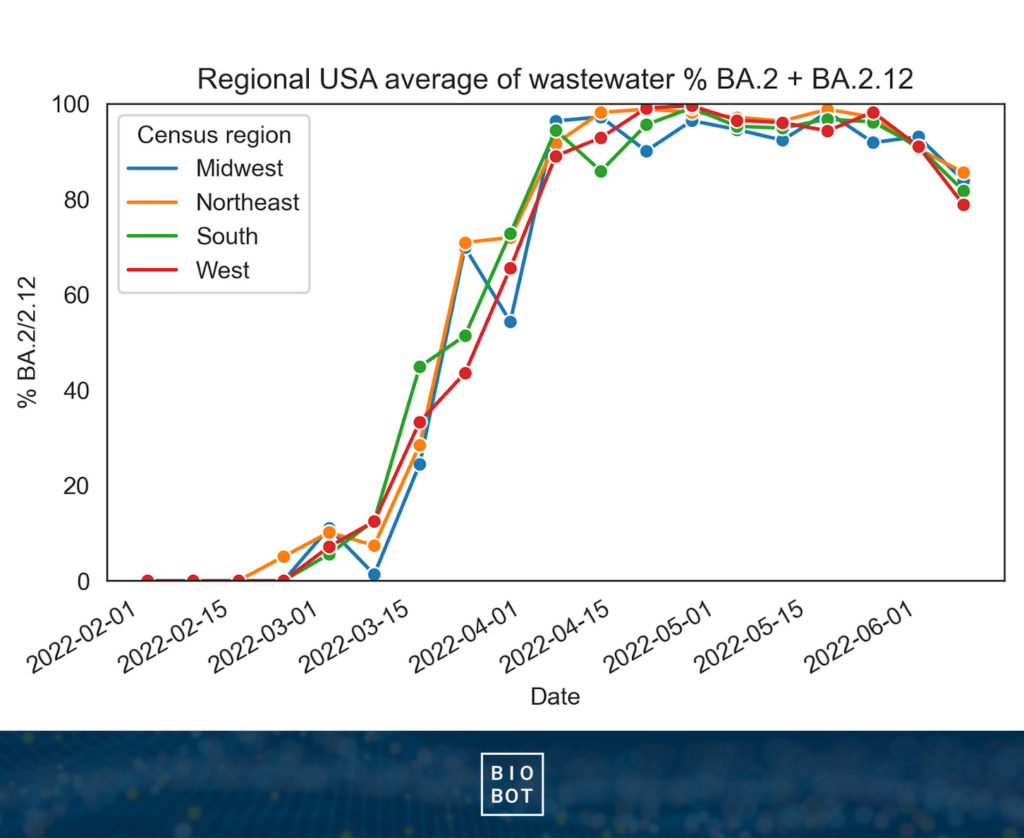
Based on our variant data, the BA.2/2.12 subvariant started climbing slowly, steadily increasing until it crested 50%, and then more quickly took over for BA.1 nationwide in late May (which had been dominant since December). BA.2/2.12 rose first in the Northeast, followed by the other regions.
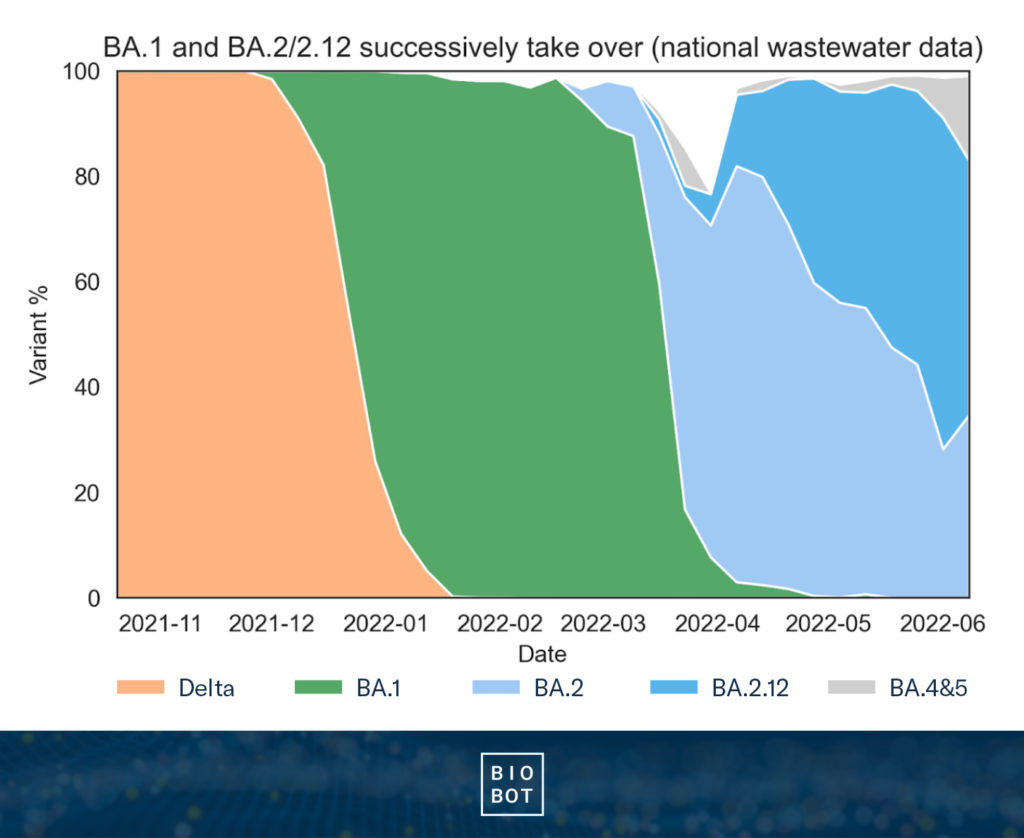
Since then, we’ve delineated between the original BA.2 and its derivative BA.2.12, and started tracking other subvariants of interest. As of early June, BA.2 and BA.2.12 combined still make up the majority of SARS-CoV-2 circulating nationwide with 34.6% and 48.4% respectively (83.0% total). BA.4&5 are at 16.0% nationwide, with the highest rates in the West (19.5%) and South (17.9%).
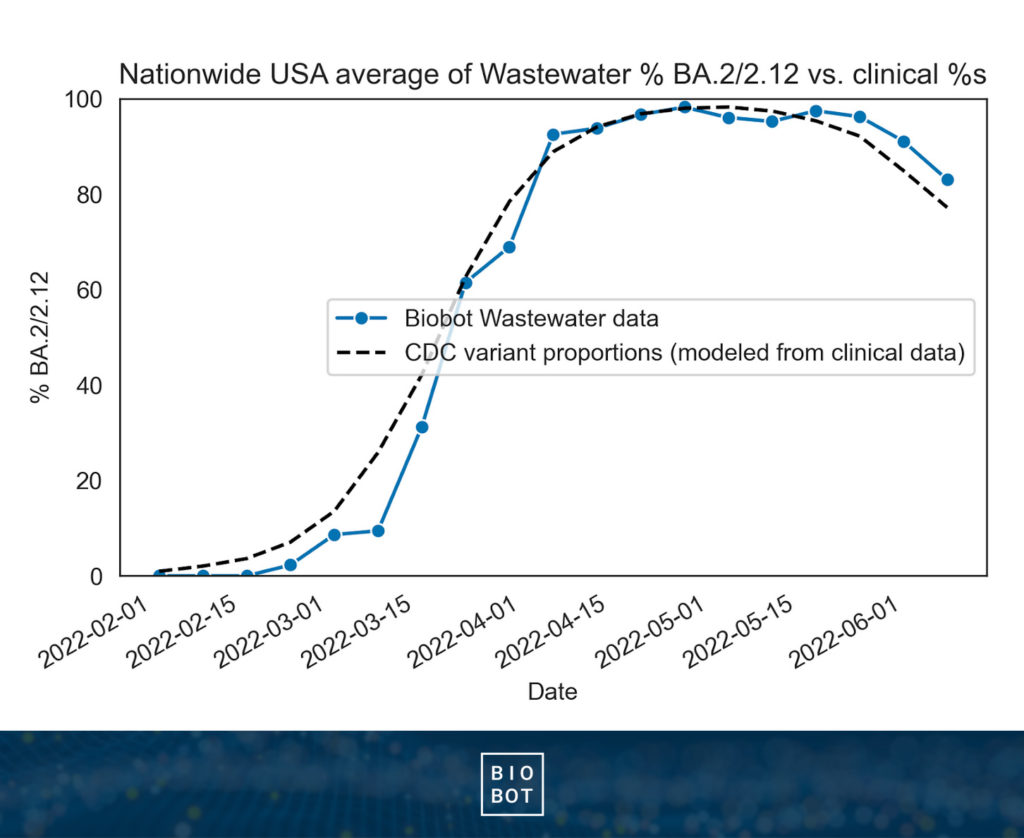
Comparing wastewater data to clinical data
At the national level, our genomic sequencing data closely mirrors the CDC’s variant proportions (which are modeled from clinical data). For smaller communities where variant tracking through clinical data can be difficult and costly, wastewater offers a cost-effective way to provide sequencing data.
Lessons for future variant monitoring
Overall, when used in concert with qPCR analysis, genomic sequencing gives us a better understanding of the SARS-CoV-2 virus. Conducting genomic sequencing on wastewater samples allows us to track the spread and trends of variants and subvariants over time. When considering how quickly new variants can evolve, arise, and sweep a community, incorporating genomic sequencing into wastewater monitoring systems enables us to stay on top of new variant outbreaks.
In addition, monitoring variants via wastewater is simple and cost-effective when compared to sequencing via clinical testing.
Lastly, we are actively monitoring the activity of Omicron subvariants, including BA.2, BA.2.12/BA 2.12.1, BA.4 and BA.5, and will regularly update our publicly available dashboard with the latest data.
Follow us on twitter for the latest trends and data analyses from our team.
Written by Biobot Analytics
Biobot provides wastewater epidemiology data & analysis to help governments & businesses focus on public health efforts and improve lives.





